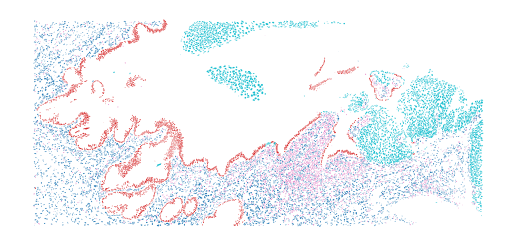
A GeoDataframe of segmented nuclei of cervical tissue crop.
Note
Pairs with: cervix_tissue_crop().
Examples:
>>> from histolytics.data import cervix_nuclei_crop
>>> ax = cervix_nuclei_crop().plot(column="class_name")
>>> ax.set_axis_off()

Source code in src/histolytics/data/fetch.py
| def cervix_nuclei_crop():
"""A GeoDataframe of segmented nuclei of cervical tissue crop.
Note:
Pairs with: `cervix_tissue_crop()`.
Examples:
>>> from histolytics.data import cervix_nuclei_crop
>>> ax = cervix_nuclei_crop().plot(column="class_name")
>>> ax.set_axis_off()
"""
return _load(BASE_PATH / "cervix_nuclei_crop.parquet")
|
